Protein Design. Week 3
Part A: Protein Analysis
Questions:
- How many molecules of amino acids do you take with a piece of 500 grams of meat? (on average an amino acid is ~100 Daltons)
meat is build out of the following proteins:
100g meat = 26 g protein (assumption)
500g meat = 130 g protein
130 g = 7.828825736E+25 daltons
7.828825736E+25 ÷ 100 = number of aa
- Why are there only 20 natural amino acids?
DNA is read in codons. The triplet of bases (nucleic acids) code for amino acid.
20 amino acids synthesise in humans. 1 amino acid relates to 2-4 codons.
In theory there could be 64 amino acids. 4(nucleic acids) to the 3 power (3-this is how many bases we need for a codon).
We must assume that we also need stop and start codons that makes 63 free possibilities for synthesising.
2 in 3 point mutatins are synonymus- this way even when there is a mistake in genetic code the outcome remains unchanged.
So this helps to have DNA translation with the highest fidelity.
Another issue is original construction of tRNA. Which is optimised for 20 amino acids.
On the molecular level:
Aminoacylation of tRNAs
ARS-tRNA recognition problem
🌿What is ARS - An aminoacyl-tRNA synthetase (aaRS or ARS), also called tRNA-ligase, is an enzyme that attaches the appropriate amino acid onto its corresponding tRNA. It does so by catalyzing the transesterification of a specific cognate amino acid or its precursor to one of all its compatible cognate tRNAs to form an aminoacyl-tRNA. In humans, the 20 different types of aa-tRNA are made by the 20 different aminoacyl-tRNA synthetases, one for each amino acid of the genetic code. This is sometimes called "charging" or "loading" the tRNA with an amino acid. Once the tRNA is charged, a ribosome can transfer the amino acid from the tRNA onto a growing peptide, according to the genetic code. Aminoacyl tRNA therefore plays an important role in RNA translation, the expression of genes to create proteins.🌿Essential amino acids. There are nine amino acids that your body can’t make. They are called essential amino acids, meaning you must have them to live. histidine, isoleucine, leucine, lysine, methionine, phenylalanine, threonine, tryptophan, and valine.🌿Anticdodon - An anticodon is a trinucleotide sequence complementary to that of a corresponding codon in a messenger RNA (mRNA) sequence.🌿One could argue that 20 is simply good enough, but several species use up to 22 residues to synthesize proteins. However, the additional two amino acids (seleno- cysteine and pyrrolysine) require alternative mechanisms for their incorporation to proteins.
Sources:
https://iubmb.onlinelibrary.wiley.com/doi/pdf/10.1080/15216540500167302
- Why most molecular helices are right handed?
The right-handed helix comes out as more stable (by about 1 kcal/mol per residue), this is not really due to either dispersion effects or entropy and must therefore arise largely from the hydrogen-bond like interactions.
- Where did amino acids come from before enzymes that make them, and before life started?
They could come from
- What do digital databases and nucleosomes have in common?
Both contain information which is somehow encoded. They are a form of organising genetic information, like databases organize digital information.
Protein
For this exercise we were asked to pick any protein (from any organism) that has a 3D structure and answer the following questions:
Again I faced a huge problem in choosing one protein.
I think Circadian Clock proteins are very inspiring. I was thinking of choosing one of them.
After all I chose...
1BET
1Bet is a protein which is a nerv growth factor protein. The organism was mouse (mus musculus). It was added in 1993. After that time more similar proteins where mapped.
- 1Bet controls the development and survival of certain neuronal populations both in the peripheral and in the central nervous systems.
- It has potential to treat Alzhaimer.
- IAmino acid sequence of my protein :
GEFSVCDSVS VWVGDKTTAT DIKGKEVTVL AEVNINNSVF RQYFFETKCR ASNPVESGCR GIDSKHWNSY CTTTHTFVKA LTTDEKQAAW RFIRIDTACV CVLSRKA
107 amino acids - long
Macromolecule Content
- Total Structure Weight: 11.99 kDa
- Atom Count: 872
- Residue Count: 107
- Unique protein chains: 1
- Does your protein belong to any protein family?
This protein belongs to the family of Neurotrophins, which guide the development of the nervous system.
Brain is composed of 85 billion interconnected neurons. Individually, each neuron receives signals from its many neighbors, and based on these signals, decides whether to dispatch its own signal to other nerve cells. Together, the combined action of all of these neurons allows us to sense the surrounding world, think about what we see, and make appropriate actions. Remarkably, this complicated structure is formed in nine short months as an embryo grows into a baby. Nerve cells start as typical, compact cells, but then they send out long axons and dendrites, connecting to other cells in the brain or even to entirely different parts of the body. Neurons in the growing brain test the connections with their neighbors, looking for the proper wiring. Half of the neurons are discarded during this process, in areas that get too crowded. The half that remain become the nervous system. Throughout the rest of life, these neurons typically do not reproduce, although they do send out more dendrites to neighboring cells as the nervous system grows or repairs damaged areas.
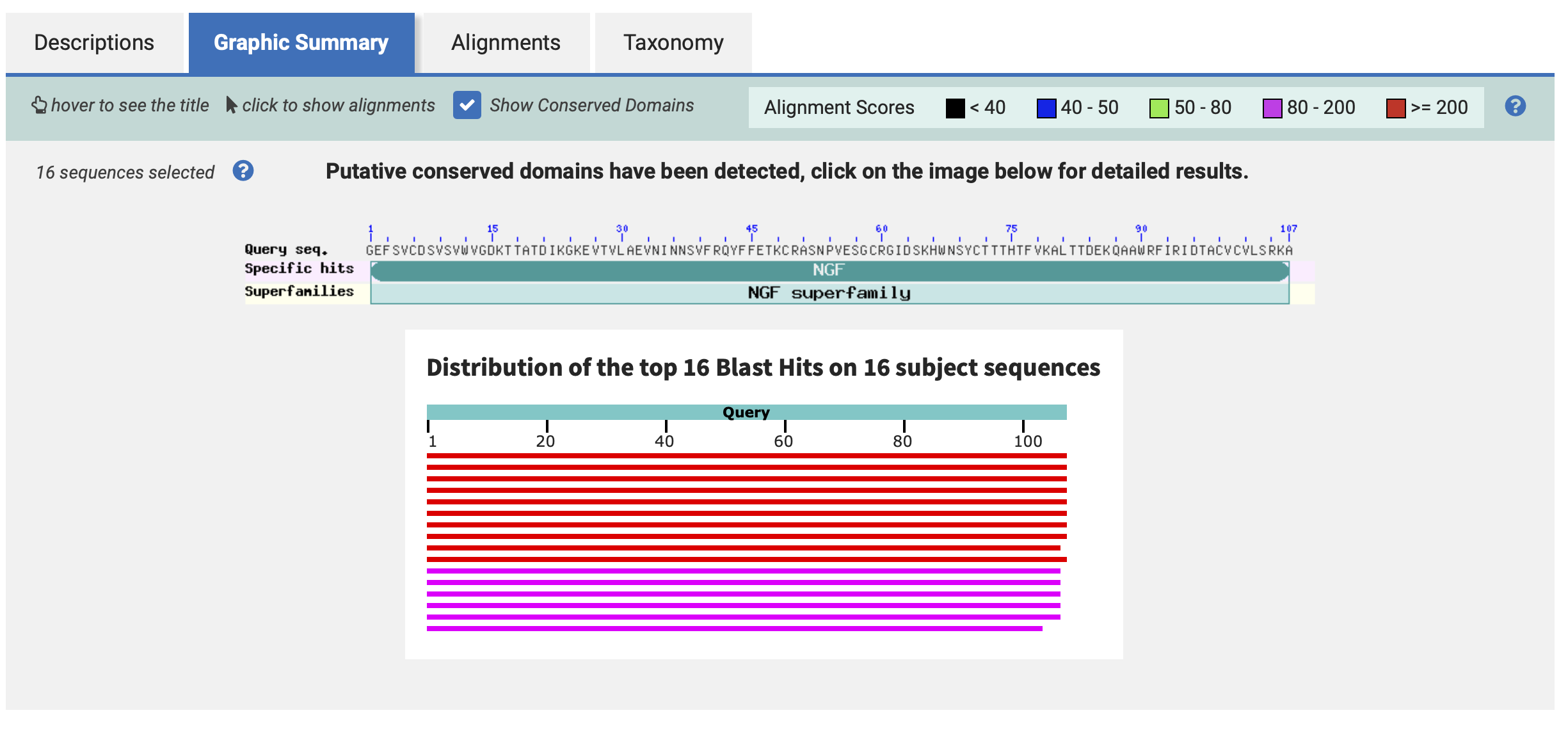
- How many protein sequence homologs are there for your protein?
Hint: Use the pBLAST tool to search for homologs and ClustalOmega to align and visualize them.
Structure page RCSB of mine protein
- Identify the structure page of your protein in RCSB
- When was the structure solved? Is it a good quality structure?
- Are there any other molecules in the solved structure apart from protein?
- Does your protein belong to any structure classification family?
- Open the structure of your protein in any 3D molecule visualization software
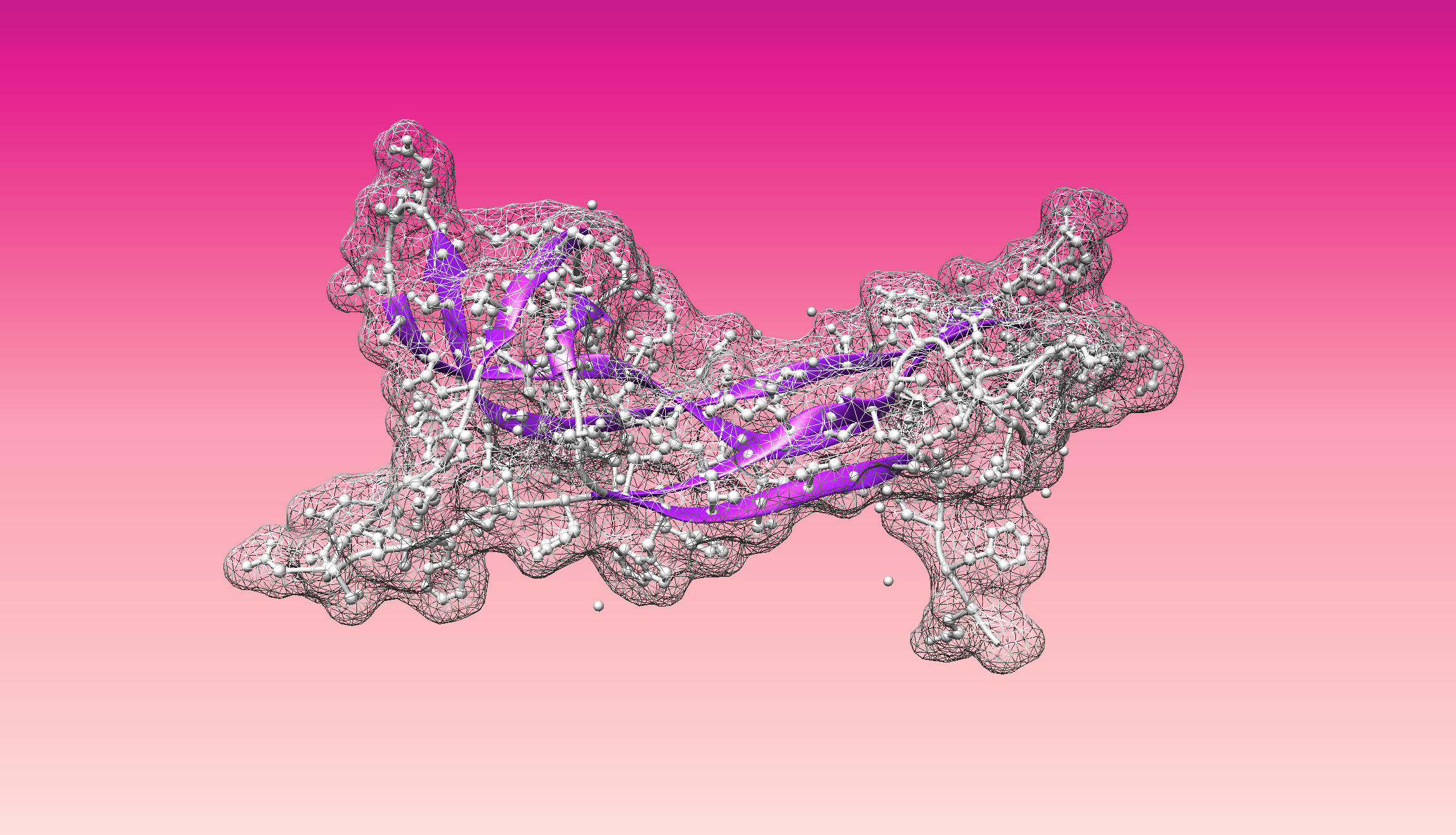
- Visualize the protein as "cartoon", "ribbon" and "ball and stick".
- Visualize the surface of the protein. Does it have any "holes" (aka binding pockets)?
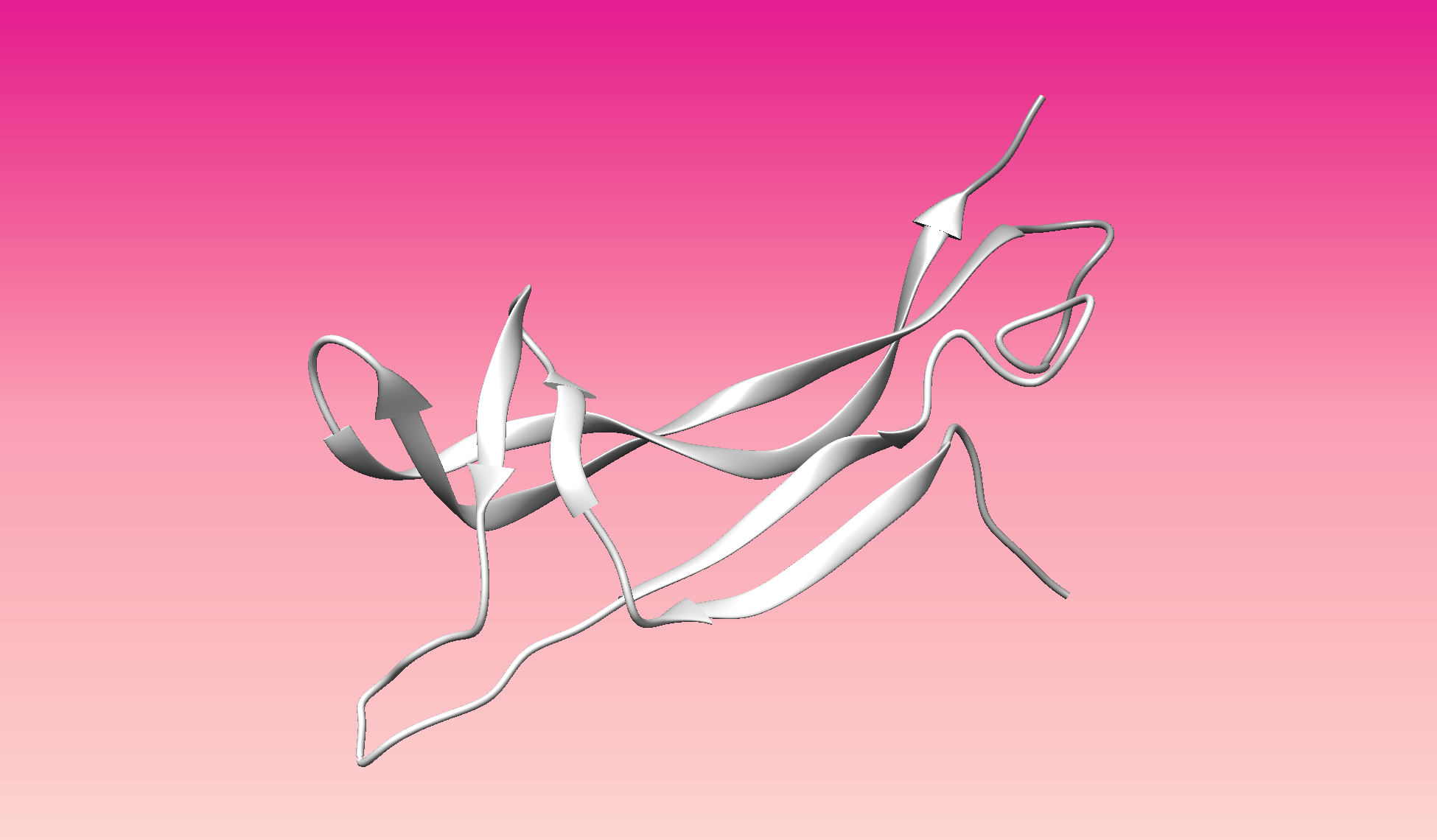
Color the protein by secondary structure. Does it have more helices or sheets?
- Color the protein by residue type. What can you tell about the distribution of hydrophobic vs hydrophilic residues?
Links:
Rubisco (http://pdb101.rcsb.org/motm/11)
PDB pioneers (http://pdb101.rcsb.org/search)
Part B: How to (almost) Fold (almost) Anything - Protein Folding
In this part you will be folding protein sequences into 3D structures. The goal is to get an understanding on how computational protein modeling works as well as to see first hand the great computing power needed for molecular simulations in biology.
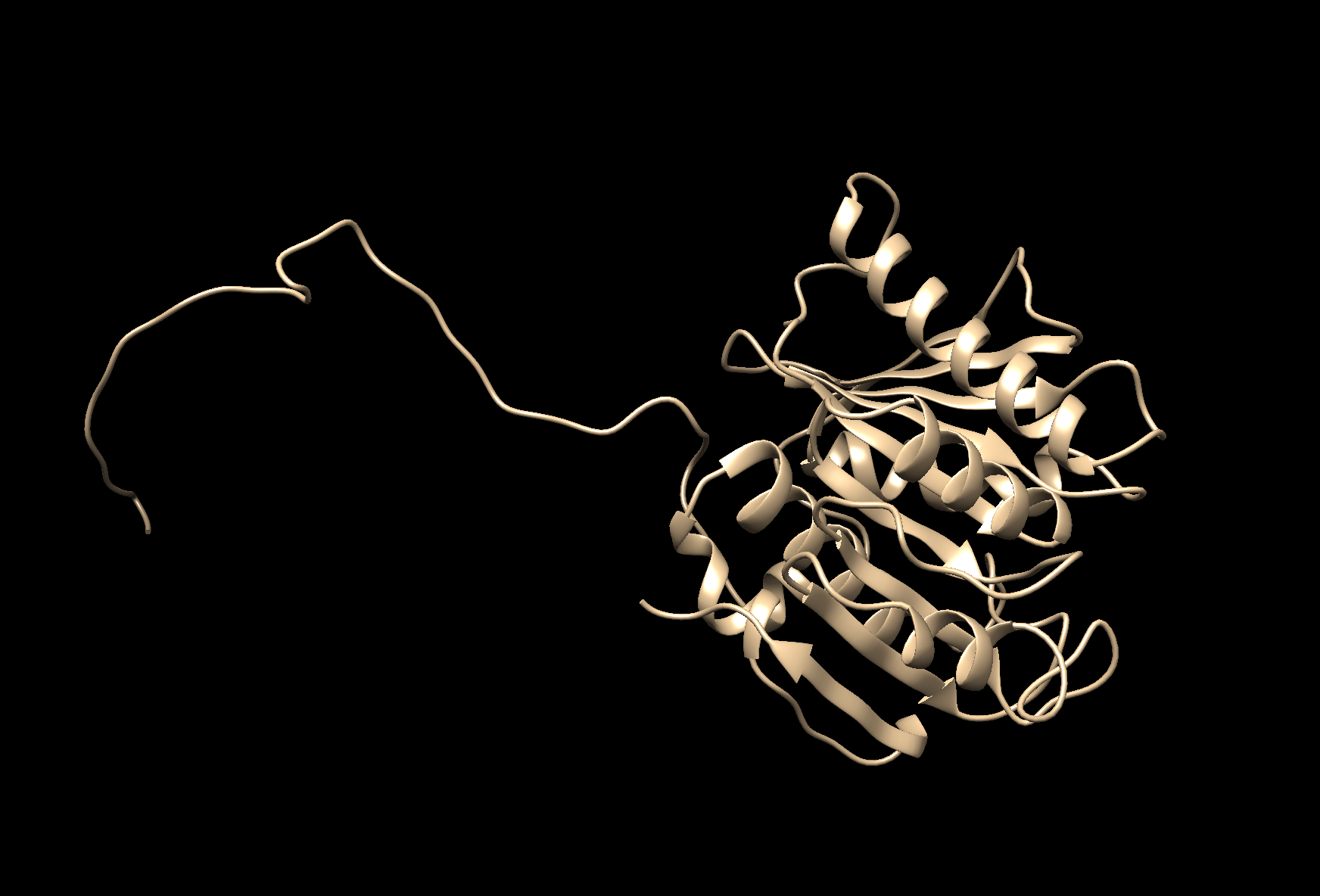
- We were asked to choose less than 100 aa protein. I
Folded Structure of the enzyme PEThase made in Robetta
3D Printed protein (unfortunately part of it was damaged during printing)
Part C: Protein Design by Machine Learning











